Projects
ProteoWizard provides a modular and extensible set of open-source, cross-platform tools and libraries. The tools perform proteomics data analyses; the libraries enable rapid tool creation by providing a robust, pluggable development framework that simplifies and unifies data file access, and performs standard chemistry and LCMS dataset computations.
The Objective
The primary goal of ProteoWizard is to eliminate the existing barriers to proteomic software development so that researchers can focus on the development of new analytic approaches, rather than having to dedicate significant resources to mundane (if important) tasks, like reading data files.
The first challenge of writing analysis tools is reading the myriad of data formats prevalent throughout the proteomics field. In addition to the open mzML, mzXML, and mzData formats, each vendor typically encodes their data in a vendor-specific, proprietary, closed format. The third challenge is the lack of a single, standard cross-platform library that performs common calculations, such as protein digestion, mass computation, peak integration, charge state detection and isotope deconvolution. Consequently, these calculations have been repeatedly implemented in the course of developing more sophisticated tools.
As the proteomics field continues to grow, an increasing number of research groups are writing custom software for data analysis. Our goal is for ProteoWizard to become a repository where proteomics software developers can share their work, and benefit from the work of others. We welcome contributions of existing code, or new projects that will benefit the proteomics community.
History
The ProteoWizard software project was initiated in 2007, by the Mallick lab, at the Spielberg Family Center for Applied Proteomics. We are extremely grateful to Dr. David Agus for his initial and ongoing support for this initiative. Since 2007, numerous other groups have been actively contributing code, and expertise to the project. Notably, the MacCoss group at University of Washington and the Tabb group at Vanderbilt Unversity. In 2010, The ProteoWizard Project was spun out from the Center for Applied Molecular Medicine, at the University of Southern California, into a California, Non-Profit Corporation - the ProteoWizard Software Foundation. We are greatly appreciative to the USC Stevens Institute for helping with the creation of the ProteoWizard Software Foundation.
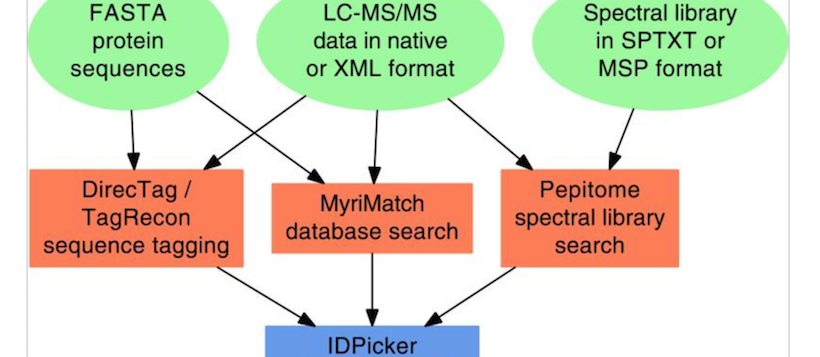
Bumbershoot is a suite of proteomic tools developed by the Tabb Lab at Vanderbilt University. The suite includes standard database search (MyriMatch), spectral library search (Pepitome), tag-based search (DirecTag/TagRecon), proteomic quality control (Quameter), and parsimonious protein assembly (IDPicker). These tools use ProteoWizard primarily for reading/writing proteomic file formats, but also for generic tasks such as in silico protein digestion.
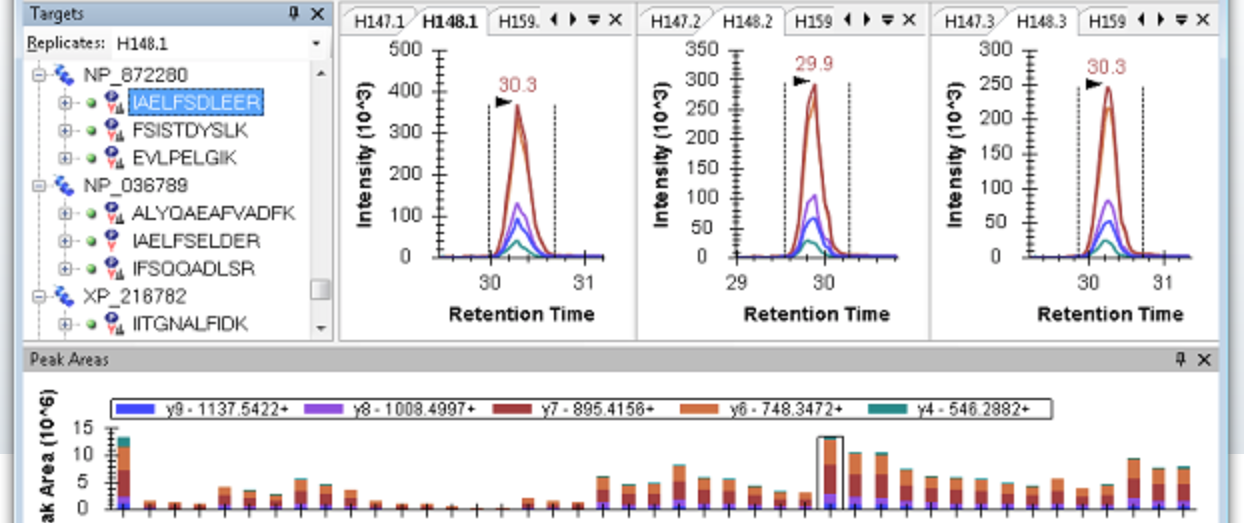
Skyline is a freely-available and open source Windows client application for building Selected Reaction Monitoring (SRM) / Multiple Reaction Monitoring (MRM), Parallel Reaction Monitoring (PRM - Targeted MS/MS), Data Independent Acquisition (DIA/SWATH) and targeted DDA with MS1 quantitative methods and analyzing the resulting mass spectrometer data. It aims to employ cutting-edge technologies for creating and iteratively refining targeted methods for large-scale quantitative mass spectrometry studies in life sciences. Skyline uses ProteoWizard to read mass spectra from either standard data formats or directly from vendor raw files.